Proteomics
Proteomics
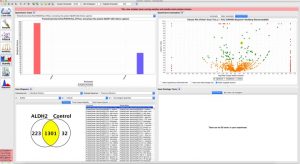
- Quantitation in proteomics has become a popular area in recent proteomics research with the development of quantitation techniques such as SILAC, TMT, and Label Free.
Stable Isotope Labeling with Amino Acids in Cell Culture
- Two populations of cells are cultivated in cell culture. One cell population is fed with growth medium containing normal amino acids while the second population is fed with growth medium containing amino acids labeled with stable isotopes (13C, 15N etc).
Isobaric tag for relative and absolute quantitation – iTRAQ and TMT- Labeling
- is a non-gel based technique used to identify and quantify proteins/peptides from different sources in one single experiment by using isotope coded covalent tags that will label the N-terminus and side chain amines of peptides from protein digestions.
Label Free Quantitation
- It has been observed the chromatographic peak areas and number of peptides observed for a protein in a LC/MS/MS run is correlated with the concentration of that particular protein.
The Mass Spectrometry Research and Education Center (MSREC) in the Department of Chemistry provides complete proteomic support from start to finish using a wide range of technology, instrumentation and bioinformatic tools. We offer a wide range of sample preparation techniques to help you extract your proteins from you sample (cells, tissue, other fluids). We offer complex sequencing from a whole cell lysate, IP pull down or other complex extract. We also offer 1D SDS-PAGE and can stain the gel with a wide range of protein stains. Proteins of interest can be sequenced for identification, confirmation, analyzed for post-translational modification, or cross-link/disulfide bond determination. The sample can be treated with a variety of protease enzymes followed by nano LC-MS/MS analysis of the peptides on a Q Exactive Orbitrap nano-LC/MS/MS. We have found this method to be sensitive and powerful, producing numerous sequences from low fmole of material.
Sample preparation is critical so please contact Dr. Basso for sample preparation guidelines. Dr Basso has 25 years of proteomic experience and is here to help you design and implement your projects.
The MSREC offers three branches of proteomics: Quantitative, Single ID and PTM analysis and Global Proteome analysis.
List of Services:
1D SDS PAGE
Albumin Removal
Phosphoproteome enrichment
Glycopeptide enrichment
Cell lysis
Protein Precipitation
TMT Labeling
Enzymatic Digestion
Post-translational modification analysis
Cross-link analysis or cysteine disufide link determination
Ultra high-resolution LC-MS intact proteins
Ultra high-resolution LC-MS/MS
de novo sequencing
SILAC or TMT Quantitation
Protein ID
Label Free Quantitation
Please see the following pdf with recommended sample preparation methods
Minimizing Contamination in Mass Spectrometry and Proteomic Experiments 2023